Research Funding
philanthropic
Background
The pan-FGFR inhibitor erda was recently FDA-approved for pretreated mUC pts harboring FGFR2/3 alterations. We explored concordance of FGFR3 mutation profiles between the primary tumor and cfDNA using the MSK-ACCESS platform. We also correlated changes in FGFR3 cfDNA mutant allele fraction (MAF) with response to drug.
Methods
Plasma samples were collected from mUC pts started on erda at baseline, on treatment (tx), and at progression. Demographic and clinical characteristics were obtained. Baseline tumors were sequenced with MSK-IMPACT (ref) and plasma samples were sequenced using MSK-ACCESS, a cfDNA platform that sequences 129 genes using unique molecular indexes to generate > 15,000x coverage and detection of somatic mutations down to 0.1% MAF.
Results
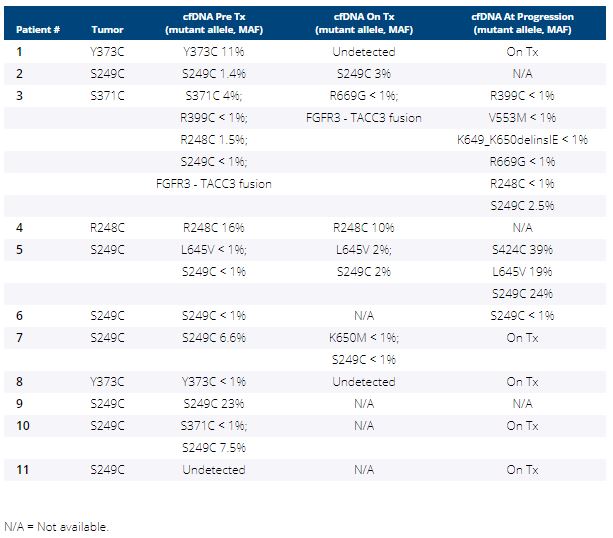
Between 08/2019-1/2020, 11 pts received erda 8mg daily. Of these, 3 pts increased dose to 9mg daily based on phosphorus level, 4 required dose reductions and 6 dose interruptions. In 5 pts, erda was discontinued for disease progression. FGFR3 S249C was the most frequent alteration detected (64%) followed by Y373C (18%), R248C and S371C (both 9%). Pre-treatment plasma FGFR3 profiles were concordant with tissue in 91% (10/11) of pts and additional FGFR3 mutations were detected in 3 cases (27%, Table), including 1 pt with an FGFR3-TACC3 fusion and hotspot mutations only in cfDNA. In 2 responding pts, the mutant allele was undetectable on erda.
Conclusions
A high degree of concordance between primary tumor and cfDNA FGFR3 mutation detection was observed. FGFR3 mutations exclusive to cfDNA were found in a subset of pts. Further pt accrual and follow-up are ongoing to assess for correlations between erda response and tx-related changes in cfDNA MAF, and to assess whether cfDNA can identify resistance mechanisms.